CBP anticorps (C-Term)
-
- Antigène Voir toutes CBP (CREBBP) Anticorps
- CBP (CREBBP) (CREB Binding Protein (CREBBP))
-
Épitope
- C-Term
-
Reactivité
- Humain
-
Hôte
- Lapin
-
Clonalité
- Polyclonal
-
Conjugué
- Cet anticorp CBP est non-conjugé
-
Application
- Western Blotting (WB), Immunofluorescence (IF), Immunoprecipitation (IP), Immunohistochemistry (Paraffin-embedded Sections) (IHC (p)), Immunocytochemistry (ICC), Chromatin Immunoprecipitation (ChIP)
- Réactivité croisée
- Humain, Singe, Souris
- Attributs du produit
-
Rabbit Polyclonal antibody to CBP (CREB binding protein)
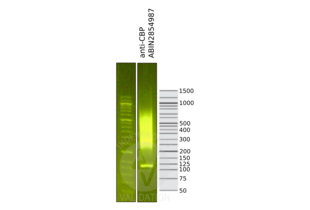
CBP antibody [C3], C-term - Purification
- Purified by antigen-affinity chromatography.
- Classe de qualité
- KO Validated
- Immunogène
- Recombinant protein encompassing a sequence within the C-terminus region of human CBP. The exact sequence is proprietary.
- Isotype
- IgG
-
-
- Indications d'application
- WB: 1:500-1:3000. ICC/IF: 1:100-1:1000. IHC-P: 1:100-1:1000. IP: 1:100-1:500. Optimal dilutions/concentrations should be determined by the researcher. Not tested in other applications.
- Commentaires
-
Positive Control: A549 , H1299 , HCT-116 , HeLa
Validation: KO/KD, Orthogonal
- Restrictions
- For Research Use only
-
- by
- Gianluca Zambanini, Anna Nordin and Claudio Cantù; Cantù Lab, Gene Regulation during Development and Disease, Linköping University
- No.
- #104437
- Date
- 07.12.2022
- Antigène
- CBP
- Numéro du lot
- 42844
- Application validée
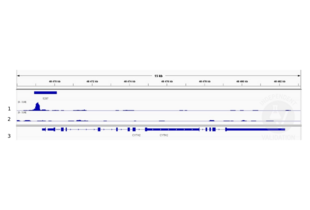
- Cleavage Under Targets and Release Using Nuclease
- Contrôle positif
Polyclonal rabbit anti-H3K4me (antibodies-online, ABIN3023251)
- Contrôle négative
Polyclonal guinea pig anti-rabbit IgG (antibodies-online, ABIN101961)
- Conclusion
Passed. ABIN2854987 allows for CUT&RUN targeted profiling of the CBP chromatin remodeler in HEK293T cells.
- Anticorps primaire
- ABIN2854987
- Anticorps secondaire
- Full Protocol
- Cell harvest and nuclear extraction
- Harvest 250,000 HEK293T cells per antibody, stimulated 24 h with 10 µM CHIR to be used at RT.
- Centrifuge cell solution 5 min at 800 x g at RT.
- Remove the liquid carefully.
- Gently resuspend cells in 1 mL of Nuclear Extraction Buffer (20 mM HEPES-KOH pH 8.2, 20% Glycerol, 0.05% IGEPAL, 0.5 mM Spermidine, 10 mM KCl, Roche Complete Protease Inhibitor EDTA-free).
- Move the solution to a 2 mL centrifuge tube.
- Pellet the nuclei 800 x g for 5 min.
- Repeat the NE wash twice for a total of three washes.
- Resuspend the nuclei in 20 µL NE Buffer per sample.
- Concanavalin A beads preparation
- Prepare one 2 mL microcentrifuge tube.
- Gently resuspend the magnetic Concanavalin A Beads (antibodies-online, ABIN6952467).
- Pipette 20 µL Con A Beads slurry for each sample into the 2 mL microcentrifuge tube.
- Place the tube on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Pipette 1 mL Binding Buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2) into the tube and resuspend ConA beads by gentle pipetting.
- Spin down the liquid from the lid with a quick pulse in a table-top centrifuge.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tube from the magnetic stand.
- Repeat the wash twice for a total of three washes.
- Gently resuspend the ConA Beads in a volume of Binding Buffer corresponding to the original volume of bead slurry, i.e. 20 µL per sample.
- Nuclei immobilization – binding to Concanavalin A beads
- Carefully vortex the nuclei suspension and add 20 µL of the Con A beads in Binding Buffer to the cell suspension for each sample.
- Close tube tightly incubates 10 min at 4 °C.
- Put the 2 mL tube on the magnet stand and when the liquid is clear remove the supernatant.
- Resuspend the beads in 1 mL of EDTA Wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, Roche Complete Protease Inhibitor EDTA-free, 2mM EDTA).
- Incubate 5 min at RT.
- Place the tube on the magnet stand and when the liquid is clear remove the supernatant.
- Resuspend the beads in 200 µl of Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, Roche Complete Protease Inhibitor EDTA-free) per sample.
- Primary antibody binding
- Divide nuclei suspension into separate 200 µL PCR tubes, one for each antibody (150,000 cells per sample).
- Add 2 µL antibody (anti-CBP antibody ABIN2854987, anti-H3K4me positive control antibody ABIN3023251, and guinea pig anti-rabbit IgG negative control antibody ABIN101961) to the respective tube, corresponding to a 1:100 dilution.
- Incubate at 4 °C ON.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Wash with 200 µL of Wash Buffer using a multichannel pipette to accelerate the process.
- Repeat the wash five times for a total of six washes.
- pAG-MNase Binding
- Prepare a 1.5 mL microcentrifuge tube containing 100 µL of pAG mix per sample (100 µL of wash buffer + 58.5 µg pAG-MNase per sample).
- Place the PCR tubes with the sample on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove tubes from the magnetic stand.
- Resuspend the beads in 100 µL of pAG-MNase premix.
- Incubate 30 min at 4 °C.
- Place the tubes on a magnet stand until the fluid is clear. Remove the liquid carefully.
- Remove the microcentrifuge tubes from the magnetic stand.
- Wash with 200 µL of Wash Buffer using a multichannel pipette to accelerate the process.
- Repeat the wash five times for a total of six washes.
- Resuspend in 100 µL of Wash Buffer.
- MNase digestion and release of pAG-MNase-antibody-chromatin complexes
- Place PCR tubes on ice and allow to chill.
- Prepare a 1.5 mL microcentrifuge tube with 102 µl of 2 mM CaCl2 mix per sample (100 µl Wash Buffer + 2 µL 100 mM CaCl2) and let it chill on ice.
- Always in ice, place the samples on the magnetic rack and when the liquid is clear remove the supernatant.
- Resuspend the samples in 100 µl of the 2 mM CaCl2 mix and incubate in ice for exactly 30 min.
- Place the sample on the magnet stand and when the liquid is clear remove the supernatant.
- Resuspend the sample in 50 µl of 1x Urea STOP Buffer (8.5 M Urea, 100 mM NaCl, 2 mM EGTA, 2 mM EDTA, 0.5% IGEPAL).
- Incubate the samples 1h at 4°C.
- Transfer the supernatant containing the pAG-MNase-bound digested chromatin fragments to fresh 200 µl PCR tubes.
- DNA Clean up
- Take the Mag-Bind® TotalPure NGS beads (Omega Bio-Tek, M1378-01) from the storage and wait until they are at RT.
- Add 2x volume of beads to each sample (e.g. 100 µL of beads for 50 µL of sample).
- Incubate the beads and the sample for 15 min at RT.
- During incubation prepare fresh EtOH 80%.
- Place the PCR tubes on a magnet stand and when the liquid is clear remove the supernatant.
- Add 200 µl of fresh 80% EtOH to the sample without disturbing the beads (Important!!! Do NOT resuspend the beads or remove the tubes from the magnet stand or the sample will be lost).
- Incubate 30 sec at RT.
- Remove the EtOH from the sample.
- Repeat the wash with 80% EtOH.
- Resuspend the beads in 25 µL of 10 mM Tris-HCl pH 8.2.
- Incubate the sample for 2 min at RT.
- Repeat the 2x beads clean up as described before (this time with 50 µL of beads for each sample).
- Resuspend the beads + DNA in 20 µL of 10 mM Tris-HCl pH 8.2.
- Library preparation and sequencing
- Prepare Libraries using KAPA HyperPrep Kit using KAPA Dual-Indexed adapters according to protocol.
- Sequence samples on an Illumina NextSeq 500 sequencer, using a NextSeq 500/550 High Output Kit v2.5 (75 Cycles), 36 bp PE.
- Peak calling
- Trim reads using using bbTools bbduk (BBMap - Bushnell B. - sourceforge.net/projects/bbmap/) to remove adapters, artifacts and repeat sequences.
- Map aligned reads to the hg38 human genome using bowtie with options -m 1 -v 0 -I 0 -X 500.
- Use SAMtools to convert SAM files to BAM files and remove duplicates.
- Use BEDtools genomecov to produce Bedgraph files.
- Call peaks using SEACR with a 0.001 threshold and the option norm stringent.
- Notes
The protocol is published in Zambanini, G. et al. A New CUT&RUN Low Volume-Urea (LoV-U) protocol uncovers Wnt/β-catenin tissue-specific genomic targets. Development (2022). PMID 36355069
Validation #104437 (Cleavage Under Targets and Release Using Nuclease)![Testé avec succès 'Independent Validation' signe]()
![Testé avec succès 'Independent Validation' signe]() Validation ImagesProtocole
Validation ImagesProtocole -
- Format
- Liquid
- Concentration
- 0.44 mg/mL
- Buffer
- 1XPBS ( pH 7), 1 % BSA, 20 % Glycerol, 0.025 % ProClin 300
- Agent conservateur
- ProClin
- Précaution d'utilisation
- This product contains ProClin: a POISONOUS AND HAZARDOUS SUBSTANCE which should be handled by trained staff only.
- Stock
- 4 °C,-20 °C
- Stockage commentaire
- Store as concentrated solution. Centrifuge briefly prior to opening vial. For short-term storage (1-2 weeks), store at 4°C. For long-term storage, aliquot and store at -20°C or below. Avoid multiple freeze-thaw cycles.
-
-
: "A new cut&run low volume-urea (LoV-U) protocol optimized for transcriptional co-factors uncovers Wnt/b-catenin tissue-specific genomic targets." dans: Development (Cambridge, England), (2022) (PubMed).
-
: "A new cut&run low volume-urea (LoV-U) protocol optimized for transcriptional co-factors uncovers Wnt/b-catenin tissue-specific genomic targets." dans: Development (Cambridge, England), (2022) (PubMed).
-
- Antigène
- CBP (CREBBP) (CREB Binding Protein (CREBBP))
- Autre désignation
- CREB binding protein (CREBBP Produits)
- Sujet
-
This gene is ubiquitously expressed and is involved in the transcriptional coactivation of many different transcription factors. First isolated as a nuclear protein that binds to cAMP-response element binding protein (CREB), this gene is now known to play critical roles in embryonic development, growth control, and homeostasis by coupling chromatin remodeling to transcription factor recognition. The protein encoded by this gene has intrinsic histone acetyltransferase activity and also acts as a scaffold to stabilize additional protein interactions with the transcription complex. This protein acetylates both histone and non-histone proteins. This protein shares regions of very high sequence similarity with protein p300 in its bromodomain, cysteine-histidine-rich regions, and histone acetyltransferase domain. Mutations in this gene cause Rubinstein-Taybi syndrome (RTS). Chromosomal translocations involving this gene have been associated with acute myeloid leukemia. Alternative splicing results in multiple transcript variants encoding different isoforms.
Cellular Localization: Nucleus - Poids moléculaire
- 265 kDa
- ID gène
- 1387
- UniProt
- Q92793
- Pathways
- TCR Signaling, Interferon-gamma Pathway, Stem Cell Maintenance, Chromatin Binding, Regulation of Lipid Metabolism by PPARalpha
-

 (1 reference)
(1 reference) (1 validation)
(1 validation)



